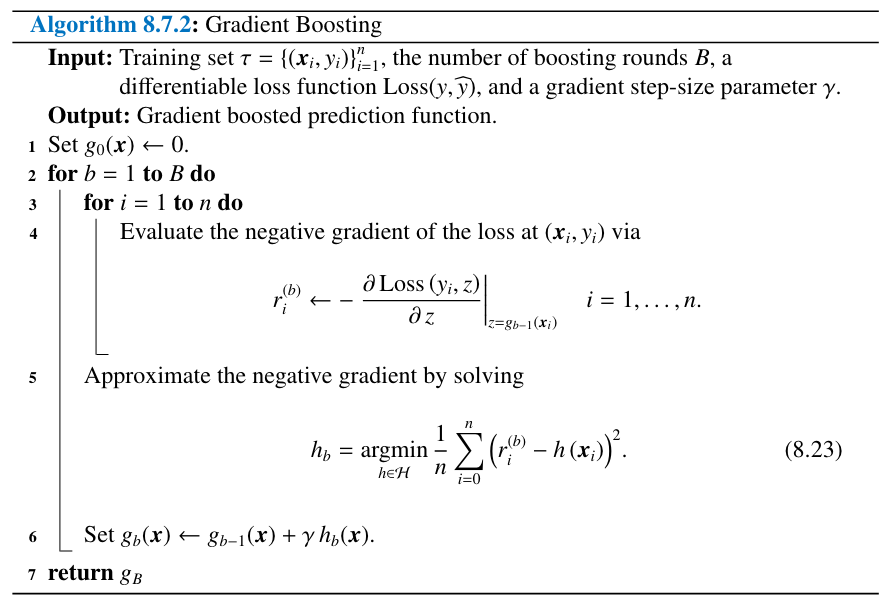
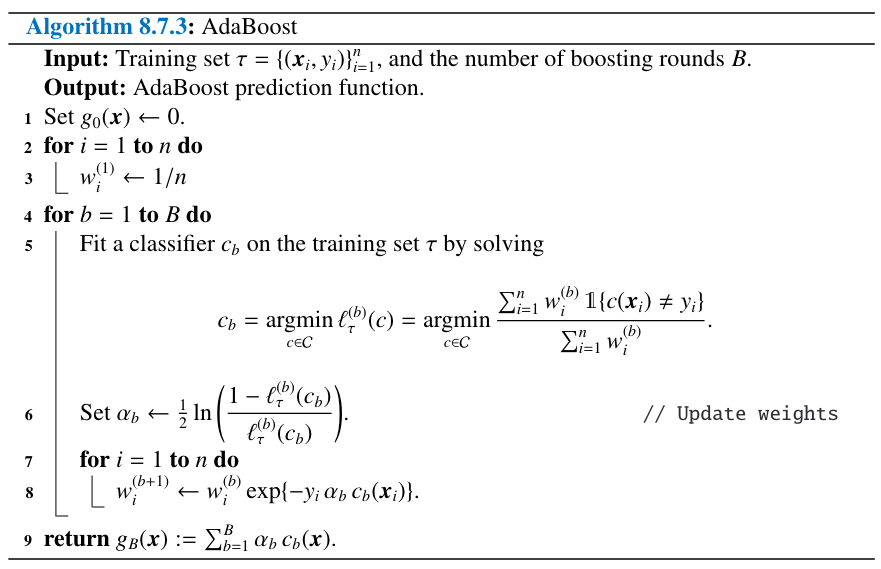
16. UNIT 8. Decision Tres#
This Unit includes main introduction to decision trees, strongly based in [1].
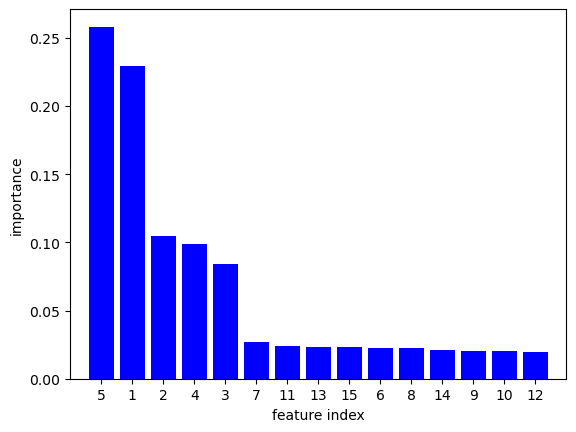
16.1. Cross-validation loss as a function of three depth#
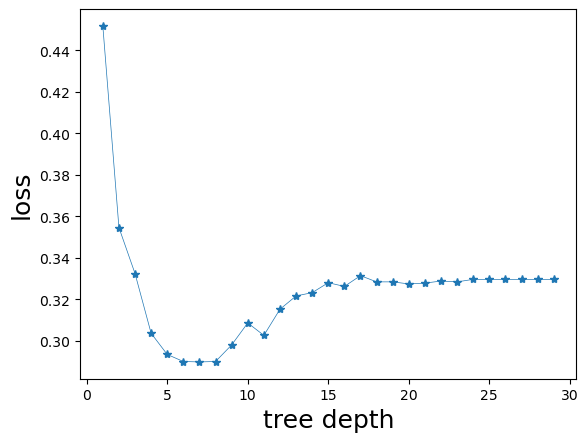
We start by seeing how sklearn builds the typical behavior of the cross-validation loss as a function of the tree depth. The cross-validation is an estimate of the expected generalization risk. Complicated trees tend to overfit the training data by producing many divisions of the feature space, as occurs with most learning methods.
""" TreeDepthCV.py """
import numpy as np
from sklearn.datasets import make_blobs
from sklearn.model_selection import cross_val_score
from sklearn.tree import DecisionTreeClassifier
from sklearn.metrics import zero_one_loss
import matplotlib.pyplot as plt
def ZeroOneScore(clf, X, y):
y_pred = clf.predict(X)
return zero_one_loss(y, y_pred)
# Construct the training set
X, y = make_blobs(n_samples=5000, n_features=10, centers=3,
random_state=10, cluster_std=10)
# construct a decision tree classifier
clf = DecisionTreeClassifier(random_state=0)
# cross validation loss as a function of tree depth (1 to 30)
xdepthlist = []
cvlist = []
tree_depth = range(1,30)
for d in tree_depth:
xdepthlist.append(d)
clf.max_depth=d
cv = np.mean(cross_val_score(clf, X, y, cv=10, scoring=ZeroOneScore))
cvlist.append(cv)
plt.xlabel('tree depth', fontsize=18, color='black')
plt.ylabel('loss', fontsize=18, color='black')
plt.plot(xdepthlist, cvlist,'-*' , linewidth=0.5)
[<matplotlib.lines.Line2D at 0x7f22c827b5d0>]
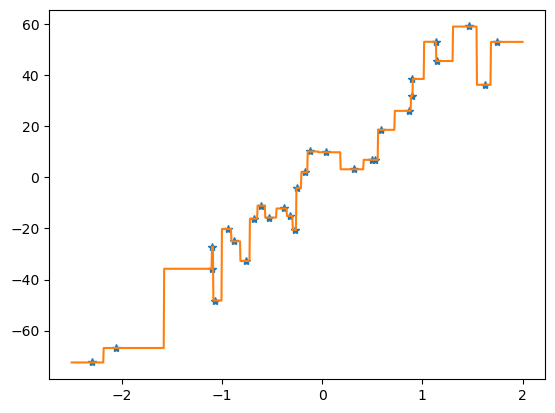
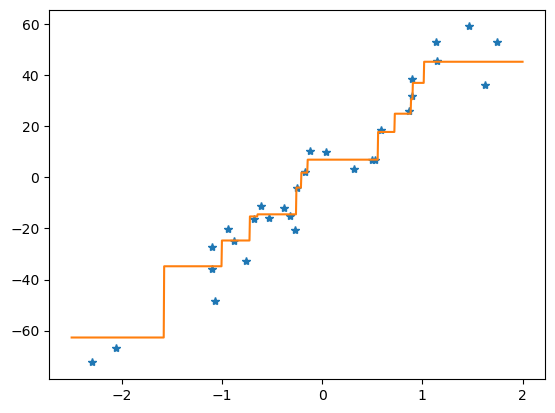
16.2. Basic step-by-step implementation#
First, we import packages and define a function to generate the training and test data
""" BasicTree.py """
import numpy as np
from sklearn.datasets import make_friedman1
from sklearn.model_selection import train_test_split
def makedata():
n_points = 500 # points
X, y = make_friedman1(n_samples=n_points, n_features=5,
noise=1.0, random_state=100)
return train_test_split(X, y, test_size=0.5, random_state=3)
now we define the main method, which calls the makedata method, uses the training data to build a regression tree and then predicts the responses, reporting the mean squared-error loss:
def main():
X_train, X_test, y_train, y_test = makedata()
maxdepth = 10 # maximum tree depth
# Create tree root at depth 0
treeRoot = TNode(0, X_train,y_train)
# Build the regression tree with maximal depth equal to max_depth
Construct_Subtree(treeRoot, maxdepth)
# Predict
y_hat = np.zeros(len(X_test))
for i in range(len(X_test)):
y_hat[i] = Predict(X_test[i],treeRoot)
MSE = np.mean(np.power(y_hat - y_test,2))
print("Basic tree: tree loss = ", MSE)
Let us specify a tree node as a Python class. Each node has a number of attributes, including features (\(X\)) and the response (\(y\)) data, as well as the depth at which the node is placed in the tree.
# tree node
class TNode:
def __init__(self, depth, X, y):
self.depth = depth
self.X = X # matrix of explanatory variables
self.y = y # vector of response variables
# initialize optimal split parameters
self.j = None
self.xi = None
# initialize children to be None
self.left = None
self.right = None
# initialize the regional predictor
self.g = None
def CalculateLoss(self):
if(len(self.y)==0):
return 0
return np.sum(np.power(self.y- self.y.mean(),2))
Implementation of the training algorithm
def Construct_Subtree(node, max_depth):
if(node.depth == max_depth or len(node.y) == 1):
node.g = node.y.mean()
else:
j, xi = CalculateOptimalSplit(node)
node.j = j
node.xi = xi
Xt, yt, Xf, yf = DataSplit(node.X, node.y, j, xi)
if(len(yt)>0):
node.left = TNode(node.depth+1,Xt,yt)
Construct_Subtree(node.left, max_depth)
if(len(yf)>0):
node.right = TNode(node.depth+1, Xf,yf)
Construct_Subtree(node.right, max_depth)
return node
to run such algorithm, we need to implement the CalculateOptinmalSplit function, and the DataSplit function in ones and zeros.
# split the data-set
def DataSplit(X,y,j,xi):
ids = X[:,j]<=xi
Xt = X[ids == True,:]
Xf = X[ids == False,:]
yt = y[ids == True]
yf = y[ids == False]
return Xt, yt, Xf, yf
def CalculateOptimalSplit(node):
X = node.X
y = node.y
best_var = 0
best_xi = X[0,best_var]
best_split_val = node.CalculateLoss()
m, n = X.shape
for j in range(0,n):
for i in range(0,m):
xi = X[i,j]
Xt, yt, Xf, yf = DataSplit(X,y,j,xi)
tmpt = TNode(0, Xt, yt)
tmpf = TNode(0, Xf, yf)
loss_t = tmpt.CalculateLoss()
loss_f = tmpf.CalculateLoss()
curr_val = loss_t + loss_f
if (curr_val < best_split_val):
best_split_val = curr_val
best_var = j
best_xi = xi
return best_var, best_xi
Finally, we implement the recursive method for prediction
def Predict(X,node):
if(node.right == None and node.left != None):
return Predict(X,node.left)
if(node.right != None and node.left == None):
return Predict(X,node.right)
if(node.right == None and node.left == None):
return node.g
else:
if(X[node.j] <= node.xi):
return Predict(X,node.left)
else:
return Predict(X,node.right)
we are now ready to run the main function providing a similar result to the sklearn implementation above
main() # run the main program
# compare with sklearn
from sklearn.tree import DecisionTreeRegressor
X_train, X_test, y_train, y_test = makedata()
regTree = DecisionTreeRegressor(max_depth = 10, random_state=0)
regTree.fit(X_train,y_train)
y_hat = regTree.predict(X_test)
MSE2 = np.mean(np.power(y_hat - y_test,2))
print("DecisionTreeRegressor: tree loss = ", MSE2)
Basic tree: tree loss = 9.067077996170276
DecisionTreeRegressor: tree loss = 10.197991295531748
16.3. Bootstrap Aggregation or bagging#
Basic bagging implementation for a regression tree, in which we compare the decision tree estimator with the corresponding bagged estimator. We use the \(R^2\) metric as a comparison.
""" BaggingExample """
import numpy as np
from sklearn.datasets import make_friedman1
from sklearn.tree import DecisionTreeRegressor
from sklearn.model_selection import train_test_split
from sklearn.metrics import r2_score
np.random.seed(100)
# create regression problem
n_points = 1000 # points
x, y = make_friedman1(n_samples=n_points, n_features=15,
noise=1.0, random_state=100)
# split to train/test set
x_train, x_test, y_train, y_test = \
train_test_split(x, y, test_size=0.33, random_state=100)
# training
regTree = DecisionTreeRegressor(random_state=100)
regTree.fit(x_train,y_train)
# test
yhat = regTree.predict(x_test)
# Bagging construction
n_estimators=500
bag = np.empty((n_estimators), dtype=object)
bootstrap_ds_arr = np.empty((n_estimators), dtype=object)
for i in range(n_estimators):
# sample bootsraped dataset
ids = np.random.choice(range(0,len(x_train)),size=len(x_train), replace=True)
x_boot = x_train[ids]
y_boot = y_train[ids]
bootstrap_ds_arr[i] = np.unique(ids)
bag[i] = DecisionTreeRegressor()
bag[i].fit(x_boot,y_boot)
# bagging prediction
yhatbag = np.zeros(len(y_test))
for i in range(n_estimators):
yhatbag = yhatbag + bag[i].predict(x_test)
yhatbag = yhatbag/n_estimators
# out of bag loss estimation
oob_pred_arr = np.zeros(len(x_train))
for i in range(len(x_train)):
x = x_train[i].reshape(1, -1)
C = []
for b in range(n_estimators):
if(np.isin(i, bootstrap_ds_arr[b])==False):
C.append(b)
for pred in bag[C]:
oob_pred_arr[i] = oob_pred_arr[i] + (pred.predict(x)[0]/len(C))
L_oob = r2_score(y_train, oob_pred_arr)
print("DecisionTreeRegressor R^2 score = ",r2_score(y_test, yhat),
"\nBagging R^2 score = ", r2_score(y_test, yhatbag),
"\nBagging OOB R^2 score = ",L_oob)
DecisionTreeRegressor R^2 score = 0.5652585102808715
Bagging R^2 score = 0.8025977878086985
Bagging OOB R^2 score = 0.821036694265204
Dirk P. Kroese, Zdravko Botev, Thomas Taimre, and Radislav: Vaisman. Data Science and Machine Learning: Mathematical and Statistical Methods. Machine Learning & Pattern Recognition. Chapman & Hall/CRC, 2020. URL: https://acems.org.au/data-science-machine-learning-book-available-download (visited on 2023-08-15).